Quantum Biology Algorithm and Application
Quantum biology refers to utilizing quantum mechanics methods in biology and chemistry to study biological systems. Many biological systems involve the conversion of external energy into forms that are usable for storage and chemical transformations. Many enzymatic systems involve chemical reactions with the substrates. These biological processes are quantum mechanical in nature.
1. Photosynthesis
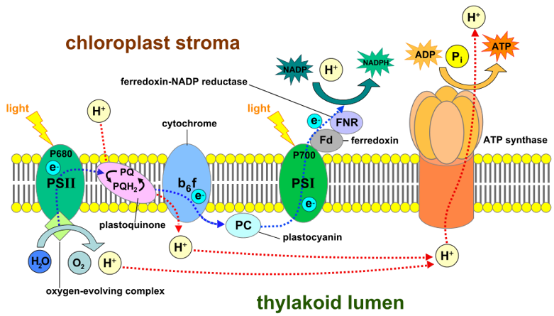
Photosynthesis is a process used by plants and other organisms to convert light energy into chemical energy stored in carbohydrate molecules, such as sugars, that can later be released to fuel the organisms' activities. The initiation of photosynthesis process is the harvest of sunlight by a network of light-absorbing molecules. The harvested energies are, then, used to produce ATP and reducing power, reduced ferredoxin, and NADPH. Last but not least, CO2 and water are captured and converted into carbohydrate molecules and other cell constituents. Since the first high-resolution structure of the Fenna-Matthews-Olson (FMO) complex was discovered 40 years ago, comprehensive understanding of the light-harvesting process occurring in photosynthetic pigment-protein complexes (PPC) has been a crucial goal. Yet, the mechanism for the energy transfer and conversion is still unclear. We explore these mechanisms through quantum biology aspect, with quantum mechanical and molecular dynamics simulation methods.
2. Non-heme enzyme
The non-heme Fe enzymes are ubiquitous in nature and perform a wide range of functions involving O2 activation. The involvement of O2 increases the difficulty of studying non-heme Fe enzyme system with quantum mechanics due to the complexity of the spin states of O2 as well as the spin crossover during the reaction. Our aim for studies in this area is to develop proper and accurate multi-spin polarizable force field for these systems. Two main features of the force field under developments involve: firstly, the force field should be able to describe multi spin states simultaneously; secondly, new methods should be adopted to calculate the switching probability in spin crossover region. Currently, force fields that fit both features are still rarely reported. In the development of the multi-spin polarizable force field, data driving model is used to build multi spin states updateable charge force filed. Machine learning methods, such as clustering and network, are employed for analyzing results and obtaining representative structures. Meanwhile, the modified Zhu-Nakamura method is introduced into molecular dynamic simulation for calculating the switching probability in spin crossover region. The strategy employed in this study is also helpful to the developments of force fields for other transition metal enzymes.
3. Polarizable Force Field in Molecular Mechanics
Molecular mechanics methods have been recognized and developed for decades, widely used for studying biomolecules. Their efficiency, as well as the developments for computing algorithms and hardware, allows the size of their subjects exceeds 1 million atoms, although the accuracy is lower comparing to the more computational demanding quantum mechanical methods. Classical molecular mechanics employs static partial atomic charge models in the energy evaluation, which ignores the polarization effect caused by the conformational and environmental changes during the biological process and simulations. Developing force fields that are capable of adjusting charges accordingly has been a major goal for recent force field development. In our studies, we utilize machine learning methods to predict atomic charges on-the-fly from the geometry of the molecule during the MD simulation. Different descriptors and mathematical algorithm for machine learning methods have been developed and tested.
High Performance Parallel Computing for Life Systems
High performance parallel computing studies parallel algorithm and software and develops parallel-capable hardware. The development of modern society and demands from scientific research require advances in computing power, therefore, high performance computing algorithms and hardware have been an essential pillar in computer science. In our group, we focus on re-constructing current algorithms in quantum biology to ensure their compatibilities for parallel computing.
Member
Jun Gao
Xiaocong Wang
Lihua Bie
